Natural Killer (NK) cells play an essential role in cancer surveillance and have a unique capability of spontaneous cytotoxicity against cancer cells. The human NK cell repertoire is functionally diversified through a tightly regulated differentiation process characterized by an early transition from CD56bright to CD56dim NK cells, followed by coordinated changes in expression of inhibitory receptors, including NKG2A and killer cell immunoglobulin-like receptors (KIR). The acquisition of self HLA class I binding KIRs during NK cell differentiation tunes the cytotoxic potential of NK cells in a process termed education, characterized by increased loading of granzyme B in dense core granules. Although NK cell differentiation and education are critical determinants of the functional potential of the cell, little is known about how these events shape the migratory behavior of NK cells.
To mediate appropriate and directed immune response against cancer, NK cells must be capable of migration to the tumor site. This process is mediated by chemokines, which guide cell migration by binding to their specific receptors. For example, in multiple myeloma, CXCR3 and CCR5 ligands (MIG, IP-10, and MIP-1a) are significantly upregulated in the bone marrow compared to healthy controls, affecting the composition of immune cells in the tumor microenvironment. In order to delineate the homing patterns of distinct NK cell subsets, we used high-dimensional flow cytometry combined with functional assays to map the NK cell chemokine receptor expression and migratory behavior.
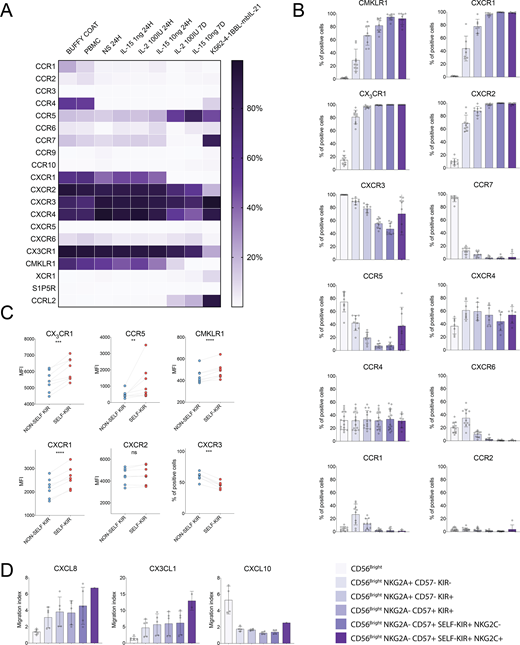
We screened resting and cytokine/feeder cell stimulated peripheral blood NK cells for the expression of a panel of 20 chemokine receptors (A). Based on CD56, CD57, NKG2A, and KIR expression, NK cells were divided into 6 phenotypically and functionally distinct subsets that were ordered according to their differentiation status (B). We found that the expression of CX3CR1, CXCR1, CXCR2, and CMKLR1 gradually increased during differentiation, whereas the expression of CXCR3, CCR7, and CCR5 was lower in more differentiated NK cells. CXCR4, CCR4, and CCR2 expression was relatively uniform across all subsets. Interestingly, CCR1 and CXCR6 were expressed mainly on less differentiated NKG2A+ CD56dim NK cells (B). Next, we stratified the chemokine receptor expression on mature KIR+ NK cells based on the expression of self (educated) or non-self KIR (uneducated). Educated NK cells expressed CXCR1, CX3CR1, CCR5, and CMKLR1 at higher levels than the uneducated NK cells. Conversely, CXCR3 was expressed at lower levels on educated NK cells (C). No difference was observed for CXCR2 expression. To determine whether the observed differences in chemokine receptor expression translate into altered chemokine responsiveness between the subsets, we combined the transwell system with multicolor flow cytometry. We found that the chemokine-induced migration capability of NK cells correlated closely with the expression level of corresponding chemokine receptor, leading to subset specific responses to various chemokine gradients (D).
The present results show that peripheral blood NK cell chemokine receptor profile changes in a coordinated fashion during NK cell differentiation and is further influenced by the expression of self-specific KIR. Interestingly, receptors which expression declines during NK cell differentiation (CCR5, CCR7, and CXCR3) are commonly associated with adaptive T cell responses to viruses, whereas receptors that are upregulated along the differentiation axis (CXCR1, CXCR2, CX3CR1, CMKLR1) are typical for neutrophils and macrophages as a part of the innate immune response. Thus, our results suggest that NK cell differentiation and education processes together shape the NK cell migratory capabilities to promote homing of the most functional NK cell subsets to the site of inflammation and serve as the first line of defense in the immune response to pathogens and tumors.
Malmberg:Fate Therapeutics: Consultancy, Patents & Royalties; Vycellix: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal